Genomic Base Editors Facilitate Artificial Evolution of Endogenous Genes in Rice
Genetic variations are very important for crop improvement. In some cases, even a single allele can make a huge difference in agricultural production. Today, many elite genes, such as wheat dwarf gene Rht, rice dwarf gene sd-1, corn haploid-inducible gene MTL, as well as herbicide-resistant genes, have been screened out from the germplasm bank and widely utilized in modern crop breeding and production. However, spontaneous mutations of genes take thousands of years in nature, and the selection efficiency is very slow and cannot meet the needs of agricultural production.
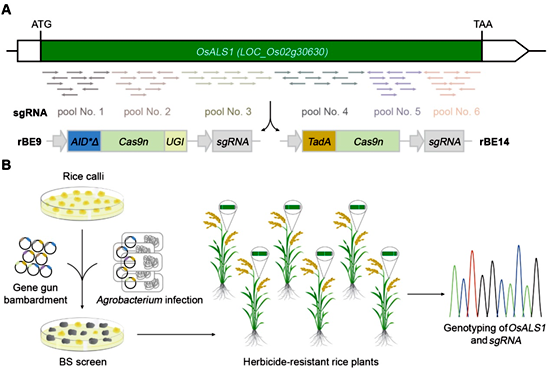
Recently, a research paper entitled “Base-Editing-Mediated Artificial Evolution of OsALS1 in Planta to Develop Novel Herbicide-tolerant Rice Germplasms”, was published in Molecular Plant by Prof. Zhou Huanbin’s group at IPP-CAAS. In this report, the authors presented the concept of artificial evolution of endogenous target genes, named Base-Editing-Mediated Gene Evolution (BEMGE/ben-ji/), which could introduce hundreds of thousands of SNPs into any target gene in rice in a short time.
In this study, a total of 63 sgRNAs were designed to cover the full-length coding region of OsALS1, it was then delivered into rice cells together with the cytosine base editors rBE9 and the adenine base editors rBE14 through both Agrobacterium-mediated and particle bombardment transformation. Deep sequencing of the OsALS1 targeted region in rice transformants showed that BEMGE generated a large number of new OsALS1 alleles. Under the selection of the herbicide bispyribac-sodium, four novel SNPs, which have been never been targeted by natural or human selection during rice domestication, were successfully identified and exhibited varying levels of tolerance to the herbicide. Furthermore, the P171F substitution identified in a strong OsALS1 allele was quickly introduced into the commercial rice cultivar Nangeng 46 through precise base editing with the corresponding base editor and sgRNA, making it a “clean-field” rice cultivar.
The proposed concept of BEMGE and its successful application in rice suggests that any endogenous gene related to a trait of interest (e.g., plant height, leaf angle, seed size, seed protein content) could be rapidly evolved for trait improvement in various crops in the near future. Thus, BEMGE opens a new era of precise molecular breeding of crops. In addition, BEMGE promotes research on gene function study given that it could dissect a protein in more details.
This study was carried out by Prof. Zhou Huanbin’ research group at IPP-CAAS in collaboration with Associate Prof. Li Shaofang at China Agricultural University, Prof. Zhou Xueping and Li Xiangju at IPP-CAAS, and Prof. Carl Spetz at Norwegian Institute of Bioeconomics.
More details are at the link below:
https://doi.org/10.1016/j.molp.2020.01.010
By Zhou Huanbin (hbzhou@ippcaas.cn)
-
 Apr 18, 2024Opening Ceremony of the Training Workshop on Wheat Head Scab Resistance Breeding and Pest Control in Africa Held in CAAS
Apr 18, 2024Opening Ceremony of the Training Workshop on Wheat Head Scab Resistance Breeding and Pest Control in Africa Held in CAAS -
 Apr 03, 2024IPPCAAS Co-organized the Training Workshop on Management and Application of Biopesticides in Nepal
Apr 03, 2024IPPCAAS Co-organized the Training Workshop on Management and Application of Biopesticides in Nepal -
 Mar 28, 2024Delegation from the School of Agriculture and Food Science of University College Dublin, Ireland Visit to IAS, CAAS
Mar 28, 2024Delegation from the School of Agriculture and Food Science of University College Dublin, Ireland Visit to IAS, CAAS -
 Mar 25, 2024Director of World Food Prize Foundation visited GSCAAS
Mar 25, 2024Director of World Food Prize Foundation visited GSCAAS -
 Mar 20, 2024Institute of Crop Sciences (ICS) and Syngenta Group Global Seeds Advance Collaborative Research in the Seed Industry
Mar 20, 2024Institute of Crop Sciences (ICS) and Syngenta Group Global Seeds Advance Collaborative Research in the Seed Industry
