分享到
A CRISPR/Cas9 Toolkit for Efficient Targeted Base Editing to Induce Genetic Variations in Rice
Gene-targeting technologies using sequence specific nucleases have been widely adopted to induce genetic modifications in plant genome, of which generating targeted point mutation for gain-of-function phenotype is of great value. Although tremendous efforts have been made in developing gene insertion or replacement approaches by harnessing DNA repair pathways through either homologous recombination or non-homologous end joining mechanisms, introducing specific DNA sequence changes into genome at the desired site remains challenging. Recently, researchers at State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection of CAAS have reported A CRISPR/Cas9 toolkit for efficient targeted base editing to induce genetic variations in rice.
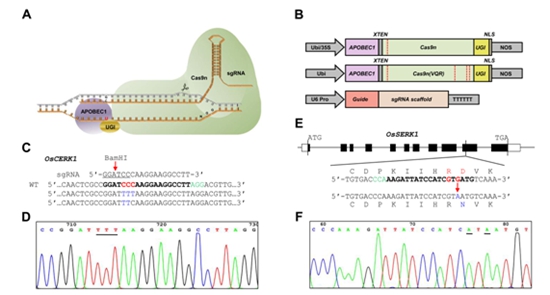
Targeted base editing in rice using the rBE3 and rBE4
This study presents an engineered Cas9 amino deaminase, rBE3, in generating gain-of-function rice mutants of OsSERK1(D428N), OsSERK2(D433N) and ipa1(S918F) genes though C > T (G > A) conversions at the target sites in rice genome with high efficiency. Furthermore, to expand the application of base editing in rice, rBE4 which recognizes different PAM sequences was also constructed and used to induce genetic variations at the key locus of rice blast susceptible gene pi-ta. Those data imply that the rBE3/rBE4 system would be a promising toolkit for generating gain-of-function mutants for fundamental molecular biology researches and materials with genetic variation for rice breeding in future.
More details are available at the bellow link:
http://engine.scichina.com/publisher/scp/journal/SCLS/doi/10.1007/s11427-016-0406-x?slug=abstract

Targeted base editing in rice using the rBE3 and rBE4
This study presents an engineered Cas9 amino deaminase, rBE3, in generating gain-of-function rice mutants of OsSERK1(D428N), OsSERK2(D433N) and ipa1(S918F) genes though C > T (G > A) conversions at the target sites in rice genome with high efficiency. Furthermore, to expand the application of base editing in rice, rBE4 which recognizes different PAM sequences was also constructed and used to induce genetic variations at the key locus of rice blast susceptible gene pi-ta. Those data imply that the rBE3/rBE4 system would be a promising toolkit for generating gain-of-function mutants for fundamental molecular biology researches and materials with genetic variation for rice breeding in future.
More details are available at the bellow link:
http://engine.scichina.com/publisher/scp/journal/SCLS/doi/10.1007/s11427-016-0406-x?slug=abstract
By Huanbin Zhou
hbzhou@ippcaas.cn
hbzhou@ippcaas.cn
Latest News
-
 Apr 18, 2024Opening Ceremony of the Training Workshop on Wheat Head Scab Resistance Breeding and Pest Control in Africa Held in CAAS
Apr 18, 2024Opening Ceremony of the Training Workshop on Wheat Head Scab Resistance Breeding and Pest Control in Africa Held in CAAS -
 Apr 03, 2024IPPCAAS Co-organized the Training Workshop on Management and Application of Biopesticides in Nepal
Apr 03, 2024IPPCAAS Co-organized the Training Workshop on Management and Application of Biopesticides in Nepal -
 Mar 28, 2024Delegation from the School of Agriculture and Food Science of University College Dublin, Ireland Visit to IAS, CAAS
Mar 28, 2024Delegation from the School of Agriculture and Food Science of University College Dublin, Ireland Visit to IAS, CAAS -
 Mar 25, 2024Director of World Food Prize Foundation visited GSCAAS
Mar 25, 2024Director of World Food Prize Foundation visited GSCAAS -
 Mar 20, 2024Institute of Crop Sciences (ICS) and Syngenta Group Global Seeds Advance Collaborative Research in the Seed Industry
Mar 20, 2024Institute of Crop Sciences (ICS) and Syngenta Group Global Seeds Advance Collaborative Research in the Seed Industry
