Arginine methylation is required for remodeling pre-mRNA splicing and induction of autophagy in rice blast fungus
Protein arginine methyltransferases (PRMTs) regulate many physiological processes, including autophagy. However, the direct roles of various PRMTs during autophagosome formation remain unclear. Recently, the research group led by Prof. Liu Wende from the Department of Plant Pathology characterized the function of MoHMT1 in the rice blast fungus, Magnaporthe oryzae. Knock out of MoHMT1 resulted in inhibited growth and a decreased ability to cause disease lesions on rice seedlings. MoHMT1 catalyzes the di-methylation of arginine 247, 251, 261, and 271 residues of MoSNP1, a U1 small nuclear RNP (snRNP) component, likely in a manner dependent on direct interaction. RNA-seq analysis revealed that alternative splicing of pre-mRNAs of 558 genes, including the autophagy-related (ATG) gene MoATG4, is altered in MoHMT1 deletion mutants, compared to wild-type strains under normal growth conditions. During light exposure or nitrogen starvation, MoHMT1 localized to autophagosomes, and MoHMT1 mutants display defects in autophagy induction. Under nitrogen-starvation, six additional MoATG genes were identified with retained introns in their mRNA transcripts, corresponding with a significant reduction in transcripts of intron-spliced isoforms in the MoHMT1 mutant strain. In conclusion, this study shows that arginine methylation plays an essential role in accurate pre-mRNA splicing necessary for a range of developmental processes, including autophagosome formation.
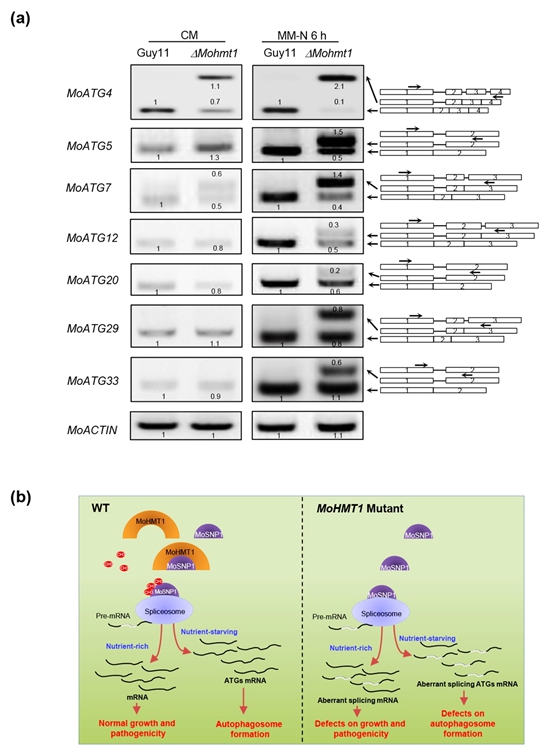
Fig. 1 A model illustrating the essential role of MoHMT1 in pre-mRNA splicing and induction of autophagy in M. oryzae. In the presence of MoHMT1 (left), MoHMT1 catalyzes the methylation of MoSNP1 in a direct binding-dependent manner. Arginine methylation of MoSNP1 regulates the proper pre-mRNA splicing that impacts diverse developmental processes, including autophagosome formation. Lack of MoHMT1-mediated arginine methylation of MoSNP1 (right), results in large scale pre-mRNA alternative splicing. In particular, under nitrogen starvation, seven autophagy-related genes generated intron retention isoforms, leading to a significant reduction in the transcript levels of intron-spliced isoforms, thus the MoHMT1-mediated remodeling MoATGs pre-mRNA splicing to compromise induction of autophagy.
By Liu Wende (wendeliu@126.com)
-
 Apr 18, 2024Opening Ceremony of the Training Workshop on Wheat Head Scab Resistance Breeding and Pest Control in Africa Held in CAAS
Apr 18, 2024Opening Ceremony of the Training Workshop on Wheat Head Scab Resistance Breeding and Pest Control in Africa Held in CAAS -
 Apr 03, 2024IPPCAAS Co-organized the Training Workshop on Management and Application of Biopesticides in Nepal
Apr 03, 2024IPPCAAS Co-organized the Training Workshop on Management and Application of Biopesticides in Nepal -
 Mar 28, 2024Delegation from the School of Agriculture and Food Science of University College Dublin, Ireland Visit to IAS, CAAS
Mar 28, 2024Delegation from the School of Agriculture and Food Science of University College Dublin, Ireland Visit to IAS, CAAS -
 Mar 25, 2024Director of World Food Prize Foundation visited GSCAAS
Mar 25, 2024Director of World Food Prize Foundation visited GSCAAS -
 Mar 20, 2024Institute of Crop Sciences (ICS) and Syngenta Group Global Seeds Advance Collaborative Research in the Seed Industry
Mar 20, 2024Institute of Crop Sciences (ICS) and Syngenta Group Global Seeds Advance Collaborative Research in the Seed Industry
