Production of novel beneficial alleles of a rice yield-related QTL by CRISPR/Cas9
Breeding high -yield crop cultivars is an effective way to improve agronomic performance for key grain crops. Traditional breeding takes years since it mainly depends on natural variation and breeder’s knowledge. Kindled by molecular technique and increased amount of genomic information, genetic dissection of beneficial trait-associated genes in elite cultivar was proved to be a good induction for future predictable breeding. Recently, in a paper entitled “Production of novel beneficial alleles of a rice yield-related QTL by CRISPR/Cas9” published on Plant Biotechnology Journal, Researchers of China National Rice Research Institute (CNRRI)reported that production of novel beneficial alleles of a rice yield-related QTL counld optimize culm strength and panicle size d. In this study, three quantitative trait loci (QTLs) associated with stem cross section area (SCSA) were firstly detected by using 127 chromosome substitution segments lines (CSSLs) derived from the offspring of 9311 crossed with Nipponbare and previous bin map made by these researchers. Fine-mapping and genomic complementary confirmed that qSCSA3-1 is allelic to SCM3/OsTB1 and has pleiotropic effects.

The sequence of OsTB1 among rice cultivars NPB (japonica), Koshihikari (japonica) , 93-11 (indica), Kasalath (indica), Zhongchao123 (japonica) and Chugoku117 (indica) have polymorphisms in the 5’-flanking coding sequence (CDS). Specially, the four varieties with large SCSA both contained a TGTG insertion at +219 in the 5’ non-coding region, suggesting that this insertion might be important for OsTB1 expression, consequently affecting SCSA.
Based on this prediction, six single-guide RNAs (sgRNAs) were designed, four targeting the promoter sequence and two, sgRNA5 and 6, targeting the region proximal to the TGTG insertion in the 5’-flanking CDS.
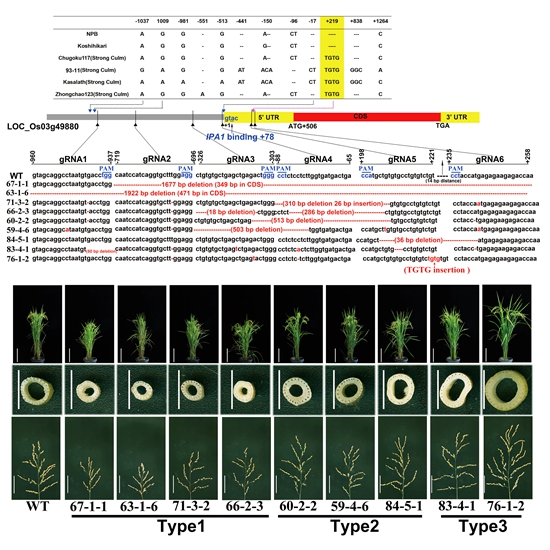
Three types of different mutations were obtained. Type 1 plants were similar to the null mutant of OsTB1, fc1-2. Type 2 plants showed similar phenotypes to the control plants. Type 3 plants showed low tillers, strong thick culm diameter and large panicle size. 76-1-2 edited-plant reproduced the same “TGTG” insertion of same site in NIP background, which is also identified an essential insertion mutation in 9311, Kasalath, ZH123 and Chokogu117 that leads to significant upregulation of OsTB1 during inflorescence development and phenotypic consequence of thick stem and large panicle size.
The research was supported by the Natural Science Foundation of China (Grant No: 31461143014). More details can be found in the link below:
https://onlinelibrary.wiley.com/doi/10.1111/PBI.13370
By Cui Yongtao(cuiyongtao20@163.com)
-
 Apr 18, 2024Opening Ceremony of the Training Workshop on Wheat Head Scab Resistance Breeding and Pest Control in Africa Held in CAAS
Apr 18, 2024Opening Ceremony of the Training Workshop on Wheat Head Scab Resistance Breeding and Pest Control in Africa Held in CAAS -
 Apr 03, 2024IPPCAAS Co-organized the Training Workshop on Management and Application of Biopesticides in Nepal
Apr 03, 2024IPPCAAS Co-organized the Training Workshop on Management and Application of Biopesticides in Nepal -
 Mar 28, 2024Delegation from the School of Agriculture and Food Science of University College Dublin, Ireland Visit to IAS, CAAS
Mar 28, 2024Delegation from the School of Agriculture and Food Science of University College Dublin, Ireland Visit to IAS, CAAS -
 Mar 25, 2024Director of World Food Prize Foundation visited GSCAAS
Mar 25, 2024Director of World Food Prize Foundation visited GSCAAS -
 Mar 20, 2024Institute of Crop Sciences (ICS) and Syngenta Group Global Seeds Advance Collaborative Research in the Seed Industry
Mar 20, 2024Institute of Crop Sciences (ICS) and Syngenta Group Global Seeds Advance Collaborative Research in the Seed Industry
