CAAS Discovered Key Candidate Genes and Long Non-coding RNA for Muscle Development of Two Dezhou Donkey Strains
Recently, Sheep genetics and breeding research group from Institute of Animal Sciences, CAAS, discovered key candidate genes and long non-coding RNA for muscle development of two Dezhou donkey strains. The results provide good ideas on further revealing donkey skeletal muscle developmental mechanism, improving the donkey meat production performance, carrying marker assisted selection in donkey breeding, and it also encourages more donkey research for the industry.
According to Prof. Zhang Li, donkeys are herbivores belonging to equine family. They are important sources of meat, donkey-hide gelatin, dairy products and donkey by-products, and play an important role in targeted poverty alleviation and Rural Revitalization. With the development of agricultural mechanization and transportation, the original service function of donkeys was almost lost, meat, skin, milk and biological products of donkeys get more and more attention. However, the level of donkey farming way is low, especially in donkey breeding.
Long non-coding RNA (lncRNA) has been extensively studied in many livestock but rarely in donkey. LncRNA plays an important role in the timing control of development, aging and death of livestock. Therefore, the study of donkey skeletal muscle lncRNA is of great significance for exploring donkey meat production performance.
In this study, RNA-Seq was used to perform high-throughput sequencing of skeletal muscle (longissimus dorsi and gluteus) of two Dezhou donkey strains (SanFen and WuTou). The differentially expressed lncRNAs were screened between different strains and tissues. Then candidate genes for conjoint analysis were screened based on GO and KEGG analysis. Finally, the accuracy of the sequencing data was verified by quantitative real-time PCR(qRT-PCR). Herein we identified 3,869 novel lncRNAs and 73 differentially expressed lncRNAs. Through the comparison between groups, specific expression of lncRNAs were found in different strains and muscle tissues. Importantly, we constructed the lncRNA-miRNA-mRNA interaction network and found 3 important candidate lncRNAs (MSTRG.9787.1, MSTRG.3144.1, MSTRG.9886.1) and 4 candidate genes (ACTN1, CDON, FMOD, BMPR1B). Analysis of the KEGG pathway indicated that the TGF-β signaling pathway acts a pivotal part in the growth and development of skeletal muscle in Dezhou donkey strains.
The related research results were published in Genes (IF = 3.759). Ms. Shi Tianpei and Dr. Hu Wenping are the co-first authors of this paper, and Prof. Zhang Li is the corresponding author. This research was funded by Dong’e E Jiao Co., Ltd. research and development program “Gene mining and identification of important economic traits in Dong’e black donkey ”(Grant No. 2016-YF-05).
More details are available at the link below:
http://www.mdpi.com/2073-4425/11/5/508
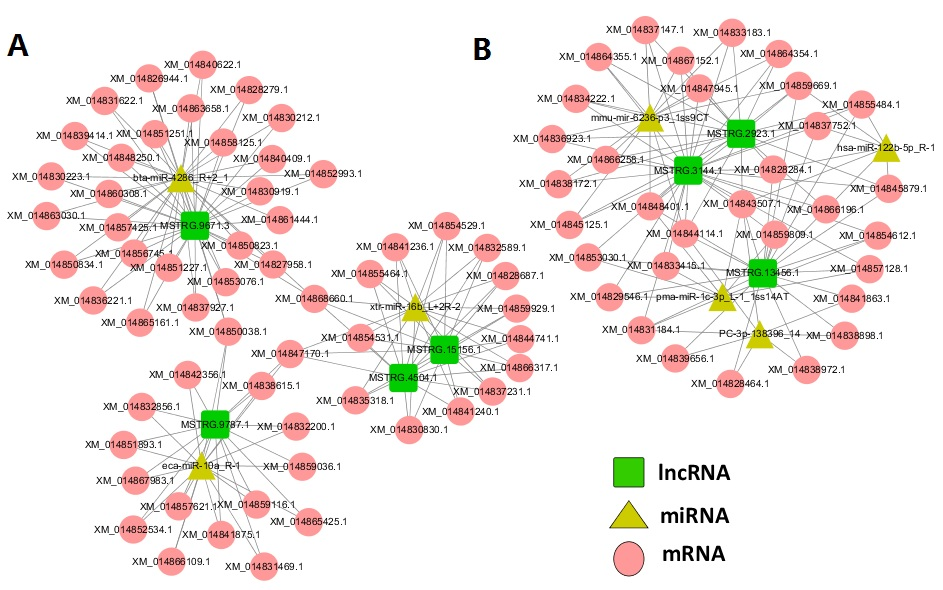
Figure 1. The ceRNA network. Green squares represent lncRNAs, yellow triangles represent genes, and red circles represent mRNAs. A: WB vs SB. B: WT vs ST. WT, WB, SB, and ST represent WuTou donkey longissimus dorsi muscle, WuTou donkey gluteus muscle, SanFen donkey longissimus dorsi muscle and SanFen donkey gluteus muscle, respectively.
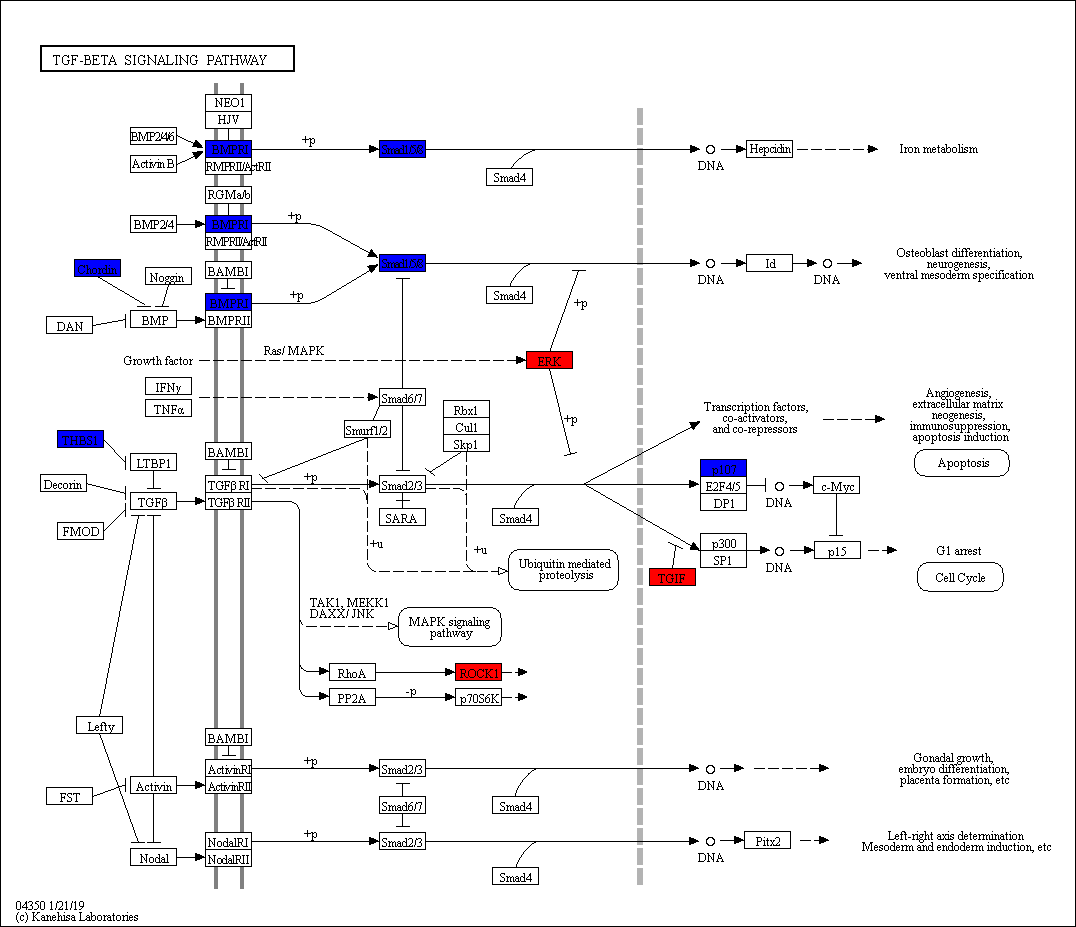
Figure 2. TGF-beta signaling pathway. Red indicates up-regulated genes and blue indicates down-regulated genes.
By Hu Wenping (huwenping@caas.cn)
-
 Apr 18, 2024Opening Ceremony of the Training Workshop on Wheat Head Scab Resistance Breeding and Pest Control in Africa Held in CAAS
Apr 18, 2024Opening Ceremony of the Training Workshop on Wheat Head Scab Resistance Breeding and Pest Control in Africa Held in CAAS -
 Apr 03, 2024IPPCAAS Co-organized the Training Workshop on Management and Application of Biopesticides in Nepal
Apr 03, 2024IPPCAAS Co-organized the Training Workshop on Management and Application of Biopesticides in Nepal -
 Mar 28, 2024Delegation from the School of Agriculture and Food Science of University College Dublin, Ireland Visit to IAS, CAAS
Mar 28, 2024Delegation from the School of Agriculture and Food Science of University College Dublin, Ireland Visit to IAS, CAAS -
 Mar 25, 2024Director of World Food Prize Foundation visited GSCAAS
Mar 25, 2024Director of World Food Prize Foundation visited GSCAAS -
 Mar 20, 2024Institute of Crop Sciences (ICS) and Syngenta Group Global Seeds Advance Collaborative Research in the Seed Industry
Mar 20, 2024Institute of Crop Sciences (ICS) and Syngenta Group Global Seeds Advance Collaborative Research in the Seed Industry
