Quantitative Proteomics Analysis Revealed Important Roles of N-glycosylation on ER Quality Control System for Development and Pathogenesis in Rice Blast Fungus
The fungal pathogen Magnaporthe oryzae can cause rice blast and wheat blast diseases, which threaten food production worldwide. During infection, M. oryzae follows a sequence of distinct developmental stages adapted to survival and invasion of the host environment. M. oryzae attaches onto the host by the conidium and then develops an appressorium to breach the host cuticle. After penetrating, it forms invasive hyphae to quickly spread within the host cells. Numerous genetic studies have focused on the mechanisms underlying every step of the infection process, but systemic approaches are needed for a broader, integrative understanding of the regulatory events during M. oryzae pathogenesis. Many infection-related signaling events are regulated through post-translational protein modifications within the pathogen. N-linked glycosylation, in which a glycan moiety is added to the amide group of an asparagine residue, is an abundant modification known to be essential for M. oryzae infection. Recently, scientists from the Department of Plant Pathology of IPP-CAAS employed a quantitative proteomics analysis to unravel the overall regulatory mechanisms of N-glycosylation at different developmental stages of M. oryzae, and the results were published online in PloS Pathogens. They detected changes in N-glycosylation levels at 559 glycosylated residues (N-glycosites) in 355 proteins during different stages and determined that the ER quality control system is elaborately regulated by N-glycosylation. Insights gained will help us better understand the regulatory mechanisms of infection in pathogenic fungi. These findings could also be important for developing novel strategies for fungal disease control.
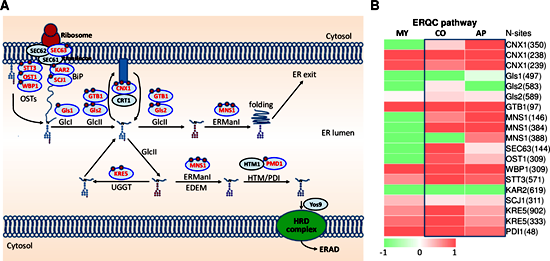
Fig. The ERQC system is regulated by N-glycosylation.
(Left) Schematic diagram of the ERQC pathway. N-glycosylated proteins were marked in red font; N-glycosites was indicated by red ring. (Right) Changes in ERQC pathway N-glycosylation protein levels in three glycosylation pathways. The colored bar represents the scale for the log10 fold change in expression from green (-2) to red (2), white is set as 0. MY, mycelia; CO, conidia; AP, appressoria.
More details can be found at the link below:
http://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1008355
By Liu Wende (wdliu@ippcaas.cn)
-
 Apr 18, 2024Opening Ceremony of the Training Workshop on Wheat Head Scab Resistance Breeding and Pest Control in Africa Held in CAAS
Apr 18, 2024Opening Ceremony of the Training Workshop on Wheat Head Scab Resistance Breeding and Pest Control in Africa Held in CAAS -
 Apr 03, 2024IPPCAAS Co-organized the Training Workshop on Management and Application of Biopesticides in Nepal
Apr 03, 2024IPPCAAS Co-organized the Training Workshop on Management and Application of Biopesticides in Nepal -
 Mar 28, 2024Delegation from the School of Agriculture and Food Science of University College Dublin, Ireland Visit to IAS, CAAS
Mar 28, 2024Delegation from the School of Agriculture and Food Science of University College Dublin, Ireland Visit to IAS, CAAS -
 Mar 25, 2024Director of World Food Prize Foundation visited GSCAAS
Mar 25, 2024Director of World Food Prize Foundation visited GSCAAS -
 Mar 20, 2024Institute of Crop Sciences (ICS) and Syngenta Group Global Seeds Advance Collaborative Research in the Seed Industry
Mar 20, 2024Institute of Crop Sciences (ICS) and Syngenta Group Global Seeds Advance Collaborative Research in the Seed Industry
